README: Bridging Medical Jargon and Lay Understanding through Expert Annotations and Data-Centric NLP
Abstract
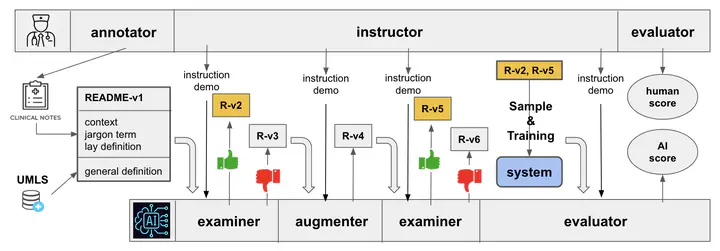
The advancement in healthcare has shifted focus toward patient-centric approaches, particularly in self-care and patient education, facilitated by access to Electronic Health Records (EHR). However, medical jargon in EHRs poses significant challenges in patient comprehension. To address this, we introduce a new task of automatically generating lay definitions, aiming to simplify complex medical terms into patient-friendly lay language. We first created the README dataset, an extensive collection of over 20,000 unique medical terms and 300,000 mentions, each offering context-aware lay definitions manually annotated by domain experts. We have also engineered a data-centric Human-AI pipeline that synergizes data filtering, augmentation, and selection to improve data quality. We then used README as the training data for models and leveraged a Retrieval-Augmented Generation (RAG) method to reduce hallucinations and improve the quality of model outputs. Our extensive automatic and human evaluations demonstrate that open-source mobile-friendly models, when fine-tuned with high-quality data, are capable of matching or even surpassing the performance of state-of-the-art closed-source large language models like ChatGPT. This research represents a significant stride in closing the knowledge gap in patient education and advancing patient-centric healthcare solutions.
Disclaimer
Submitted to ACL Rolling Review. This work is done when I was undergraduate research assistant at UMass BioNLP Lab.