Publications
publications by categories in reversed chronological order. generated by jekyll-scholar.
2024
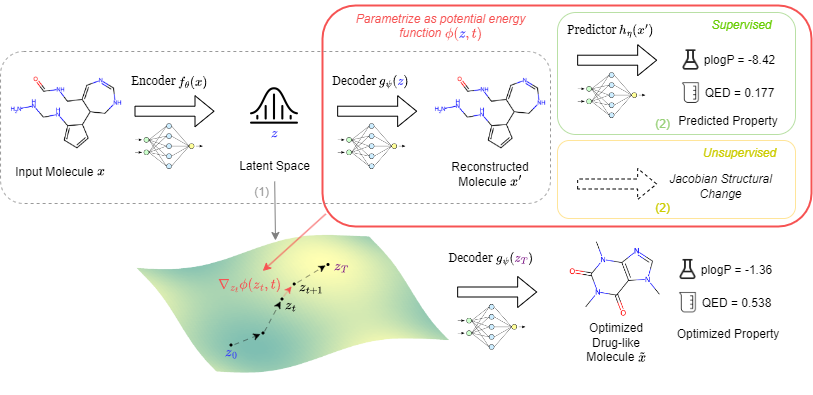
- Navigating Chemical Space with Latent FlowsIn Advances in Neural Information Processing Systems, 2024
Recent progress of deep generative models in the vision and language domain has stimulated significant interest in more structured data generation such as molecules. However, beyond generating new random molecules, efficient exploration and a comprehensive understanding of the vast chemical space are of great importance to molecular science and applications in drug design and materials discovery. In this paper, we propose a new framework, ChemFlow, to traverse chemical space through navigating the latent space learned by molecule generative models through flows. We introduce a dynamical system perspective that formulates the problem as learning a vector field that transports the mass of the molecular distribution to the region with desired molecular properties or structure diversity. Under this framework, we unify previous approaches on molecule latent space traversal and optimization and propose alternative competing methods incorporating different physical priors. We validate the efficacy of ChemFlow on molecule manipulation and single- and multi-objective molecule optimization tasks under both supervised and unsupervised molecular discovery settings. Codes and demos are publicly available on GitHub at https://github.com/garywei944/ChemFlow.
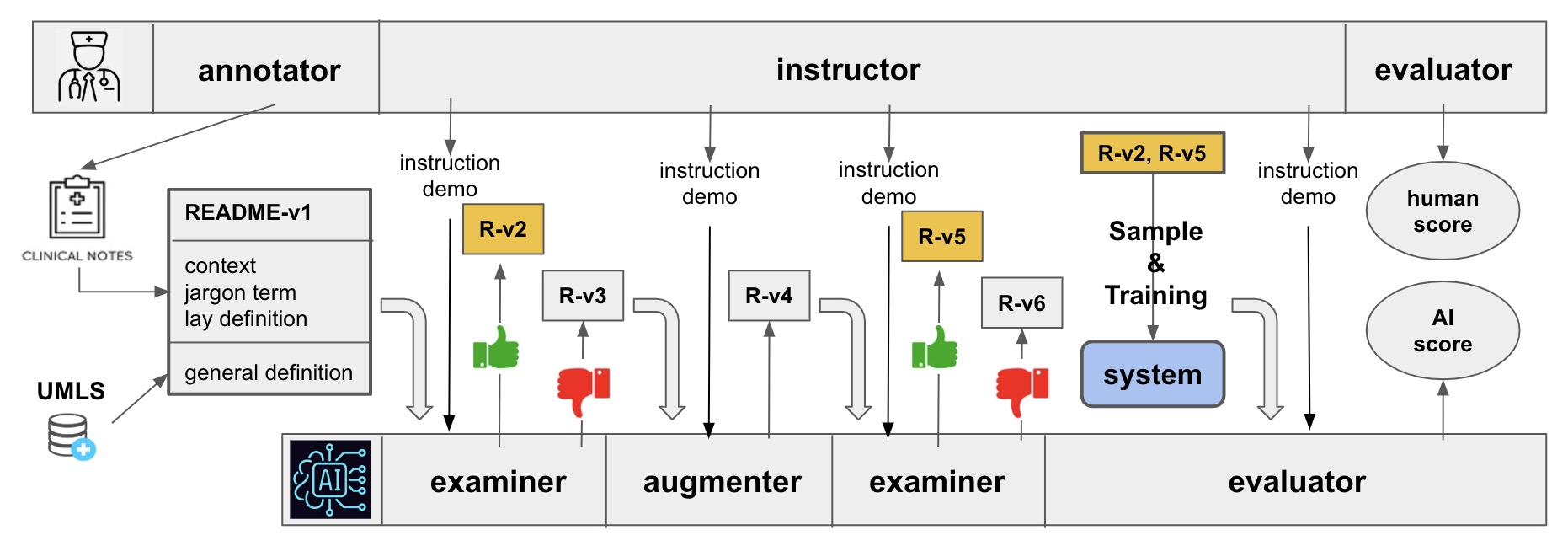
@inproceedings{wei2024navigating, title = {Navigating Chemical Space with Latent Flows}, author = {Wei, Guanghao and Huang, Yining and Duan, Chenru and Song, Yue and Du, Yuanqi}, year = {2024}, booktitle = {Advances in Neural Information Processing Systems}, editor = {Globerson, A. and Mackey, L. and Belgrave, D. and Fan, A. and Paquet, U. and Tomczak, J. and Zhang, C.}, pages = {58663--58697}, volume = {37}, publisher = {Curran Associates, Inc.}, doi = {10.52202/079017-1870}, archiveprefix = {arXiv}, primaryclass = {cs.LG}, url = {https://proceedings.neurips.cc/paper_files/paper/2024/file/6bbefb73c0ede70635823a18426b9208-Paper-Conference.pdf}, } - README: Bridging Medical Jargon and Lay Understanding for Patient Education through Data-Centric NLPZonghai Yao, Nandyala Siddharth Kantu, Guanghao Wei, Hieu Tran, Zhangqi Duan, and 4 more authorsIn Proceedings of the 2024 Conference on Empirical Methods in Natural Language Processing, 2024
The advancement in healthcare has shifted focus toward patient-centric approaches, particularly in self-care and patient education, facilitated by access to Electronic Health Records (EHR). However, medical jargon in EHRs poses significant challenges in patient comprehension. To address this, we introduce a new task of automatically generating lay definitions, aiming to simplify complex medical terms into patient-friendly lay language. We first created the README dataset, an extensive collection of over 50,000 unique (medical term, lay definition) pairs and 300,000 mentions, each offering context-aware lay definitions manually annotated by domain experts. We have also engineered a data-centric Human-AI pipeline that synergizes data filtering, augmentation, and selection to improve data quality. We then used README as the training data for models and leveraged a Retrieval-Augmented Generation method to reduce hallucinations and improve the quality of model outputs. Our extensive automatic and human evaluations demonstrate that open-source mobile-friendly models, when fine-tuned with high-quality data, are capable of matching or even surpassing the performance of state-of-the-art closed-source large language models like ChatGPT. This research represents a significant stride in closing the knowledge gap in patient education and advancing patient-centric healthcare solutions.
@inproceedings{yao-etal-2024-readme, title = {README: Bridging Medical Jargon and Lay Understanding for Patient Education through Data-Centric NLP}, author = {Yao, Zonghai and Kantu, Nandyala Siddharth and Wei, Guanghao and Tran, Hieu and Duan, Zhangqi and Kwon, Sunjae and Yang, Zhichao and annotation team, README and Yu, Hong}, year = {2024}, booktitle = {Proceedings of the 2024 Conference on Empirical Methods in Natural Language Processing}, editor = {Al-Onaizan, Yaser and Bansal, Mohit and Chen, Yun-Nung}, pages = {12609--12629}, volume = {37}, publisher = {Association for Computational Linguistics}, doi = {10.18653/v1/2024.findings-emnlp.737}, archiveprefix = {arXiv}, primaryclass = {cs.CL}, url = {https://aclanthology.org/2024.findings-emnlp.737/}, }
2023
-
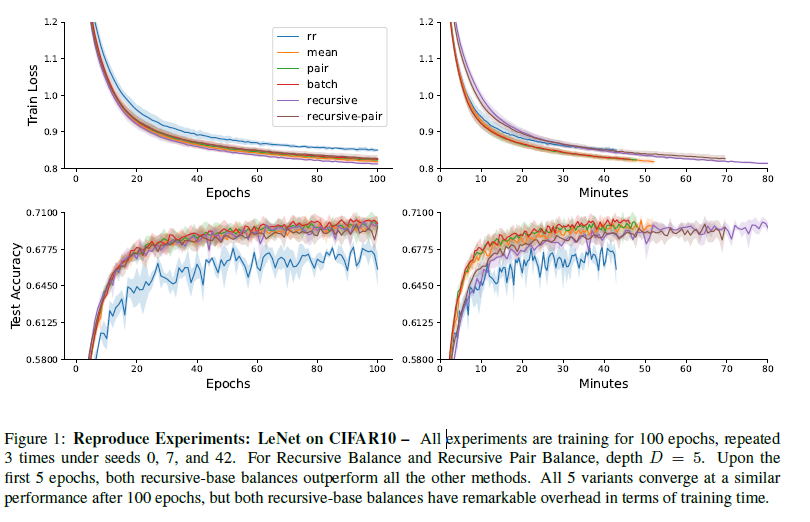 GraB-sampler: Optimal Permutation-based SGD Data Sampler for PyTorchGuanghao Wei2023
GraB-sampler: Optimal Permutation-based SGD Data Sampler for PyTorchGuanghao Wei2023The online Gradient Balancing (GraB) algorithm greedily choosing the examples ordering by solving the herding problem using per-sample gradients is proved to be the theoretically optimal solution that guarantees to outperform Random Reshuffling. However, there is currently no efficient implementation of GraB for the community to easily use it.
This work presents an efficient Python library, GraB-sampler, that allows the community to easily use GraB algorithms and proposes 5 variants of the GraB algorithm. The best performance result of the GraB-sampler reproduces the training loss and test accuracy results while only in the cost of 8.7% training time overhead and 0.85% peak GPU memory usage overhead.@misc{wei2023grabsampler, title = {GraB-sampler: Optimal Permutation-based SGD Data Sampler for PyTorch}, author = {Wei, Guanghao}, year = {2023}, doi = {10.48550/arXiv.2309.16809}, archiveprefix = {arXiv}, primaryclass = {cs.LG}, } -
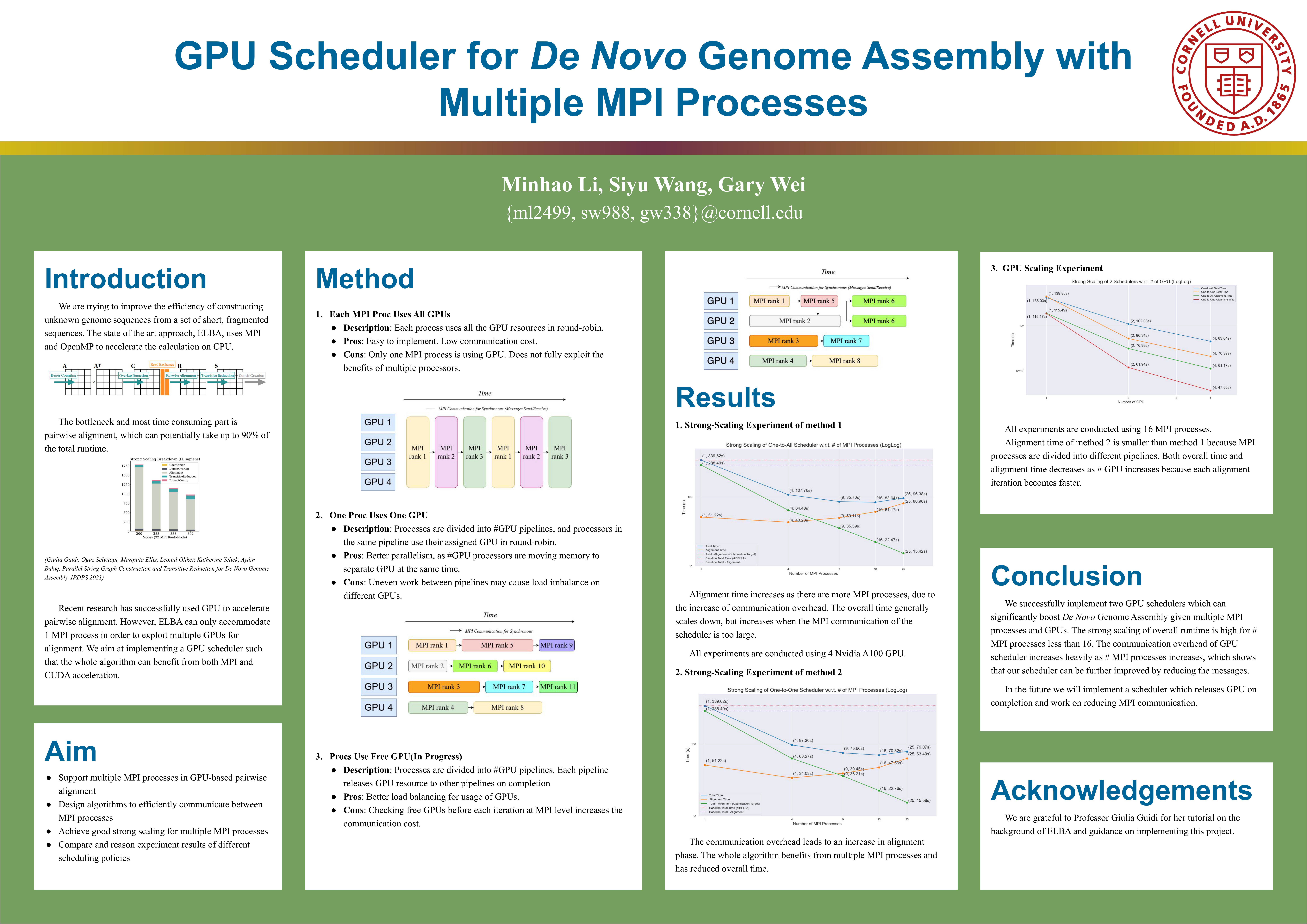 GPU Scheduler for De Novo Genome Assembly with Multiple MPI ProcessesMinhao Li*, Siyu Wang*, and Guanghao Wei*2023
GPU Scheduler for De Novo Genome Assembly with Multiple MPI ProcessesMinhao Li*, Siyu Wang*, and Guanghao Wei*2023De Novo Genome assembly is one of the most important tasks in computational biology. ELBA is the state-of-the-art distributed-memory parallel algorithm for overlap detection and layout simplification steps of DDe Novo genome assembly but exists a performance bottleneck in pairwise alignment.
In this work, we proposed 3 GPU schedulers for ELBA to accommodate multiple MPI processes and multiple GPUs. The GPU schedulers enable multiple MPI processes to perform computation on GPUs in a round-robin fashion. Both strong and weak scaling experiments show that 3 schedulers are able to significantly improve the performance of baseline while there is a trade-off between parallelism and GPU scheduler overhead. For the best performance implementation, the one-to-one scheduler achieves ∼7-8× speed-up using 25 MPI processes compared with the baseline vanilla ELBA GPU scheduler.@misc{li2023gpu, title = {GPU Scheduler for De Novo Genome Assembly with Multiple MPI Processes}, author = {Li, Minhao and Wang, Siyu and Wei, Guanghao}, year = {2023}, doi = {10.48550/arXiv.2309.07270}, archiveprefix = {arXiv}, primaryclass = {cs.DC}, }